Molecular Genetics & Immunology

Molecular genetics and immunology converge to decode how inherited variants, somatic alterations, epigenetic programs, and immune signaling networks shape disease onset, trajectory, and therapeutic response. In clinical practice, this integrated lens enables more accurate diagnostics, earlier risk stratification, and tailored interventions that align with a patient’s molecular and immunological profile. Within this paradigm, CLRD’s treatment specialization operationalizes bench‑to‑bedside advances by embedding omics‑driven testing, immune pathway mapping, and cell‑based or molecularly guided therapies into routine care for inflammatory, infectious, autoimmune, metabolic, oncologic, and neurovascular conditions.
Genetic Markers: Foundations for Precision Diagnosis and Risk Prediction
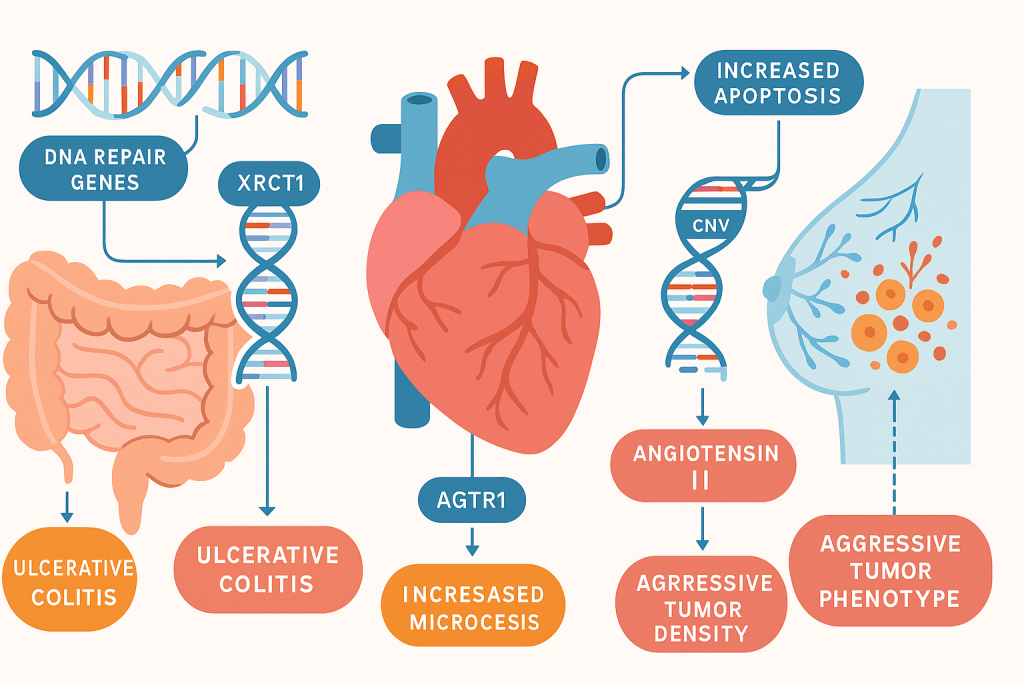
Genetic markers, single nucleotide polymorphisms, haplotypes, copy‑number variations, and regulatory sequence changes, serve as objective signals to localize disease mechanisms and predict clinical outcomes. For inflammatory bowel disease, functional polymorphisms in DNA repair genes such as XRCC1 and APE1 have been associated with heightened apoptosis and increased ulcerative colitis risk, reinforcing the role of genome integrity pathways in mucosal resilience and inflammation control. CLRD’s prior haplotype analyses of DNA repair gene polymorphisms further demonstrated significant associations with ulcerative colitis, enabling translation into a genotyping panel for prognosis and therapy planning. In cardiometabolic disease, angiotensin II type‑1 receptor gene variants correlated with circulating angiotensin II levels in essential hypertension, providing a molecular lever for anti‑hypertensive selection and dose optimization. Across oncology, promoter methylation markers such as LCN2 have been linked to microvessel density and aggressive breast tumor phenotypes, guiding decisions around anti‑angiogenic strategies and surveillance intensity.
Epigenetics and Transcriptomics: Immune Pathway Readouts in Real Time
Beyond static DNA variation, dynamic epigenetic states and transcriptional programs provide a real‑time window into immune activation, stress responses, and cytokine output. Meta‑analyses of transcriptomes in rheumatoid arthritis and ulcerative colitis identified reproducible immune signatures enriched for RNA‑binding proteins that orchestrate post‑transcriptional control of inflammatory mediators, an insight that underpins CLRD’s adoption of signature‑level diagnostics for monitoring disease activity and drug efficacy. Mechanistic studies revealed that amino‑acid starvation and integrated stress responses dampen IL‑1β production via riboclustering and autophagy, clarifying why nutritional or metabolic interventions can modulate inflammasome‑related diseases and why stress‑response pathways are attractive therapeutic targets.
Innate and Adaptive Immunity: From Pathogenesis to Therapeutic Targeting
Innate immune pattern recognition and complement lectin pathways set the stage for downstream adaptive responses. Variants in the MBL‑2 gene and corresponding serum mannan‑binding lectin levels have been associated with ulcerative colitis and Crohn’s disease, underscoring complement pathway contributions to mucosal defense and dysregulated inflammation. In infectious and sepsis settings, mitochondrial damage‑associated molecular patterns, including circulating mitochondrial DNA, amplify systemic inflammation and correlate with organ dysfunction, establishing actionable biomarkers for risk stratification and escalation of care. Endothelial dysfunction markers such as soluble thrombomodulin and soluble endoglin further refine prognostication in surgical sepsis and guide endothelial‑targeted supportive therapies.
CLRD Diagnostic Capability: Omics‑Anchored Workflows and Immune Monitoring
CLRD integrates genotyping, epigenetic profiling, transcriptome meta‑signatures, and cell‑free nucleic acid quantification into a unified diagnostic pathway. The laboratory’s polymorphism panels for XRCC1, APE1, CD14, and macrophage migration inhibitory factor are paired with inflammatory microRNA expression analysis to delineate drivers of ankylosing spondylitis and polyarthralgia, directly informing selection of TNF‑α, IL‑17, or JAK pathway blockade. Circulating microRNA‑21 assays support non‑invasive detection and risk assessment in prostate cancer and pulmonary fibrosis post‑COVID‑19, enabling cross‑tissue surveillance of fibrotic remodeling and oncogenic signaling. For neurovascular disease, CLRD’s cell‑free DNA workflows quantify mitochondrial and nuclear cfDNA fractions from a single plasma sample, with demonstrations in ischemic stroke patients that support longitudinal monitoring of hypoxia‑driven injury and secondary complications. In critical care, the center’s sepsis panels measure neutrophil extracellular trap surrogates and mitochondrial DNA release, translating molecular alarms into triage and therapy escalation protocols.
Treatment Specialization: Personalized Therapeutic Pathways
CLRD’s treatment specialization is anchored in matching molecular drivers to targeted interventions. In inflammatory bowel disease, patients harboring high‑risk DNA repair polymorphisms or elevated mucosal oxidative signatures are triaged to antioxidant adjuncts, mucosal healing regimens, and early biologic initiation, with pharmacodynamic monitoring via transcriptomic and microRNA markers to detect early non‑response and pivot therapy. Autoimmune and autoinflammatory conditions benefit from pathway‑specific selection: complement lectin deficiencies prompt tailored prophylaxis and microbiome‑conscious strategies; integrated stress response activation patterns justify use of agents that restore proteostasis or modulate autophagy; and IL‑1β‑centric signatures inform inflammasome inhibitors and dietary interventions that stabilize amino‑acid and metabolic flux. In oncology, epigenetic markers such as LCN2 promoter methylation and claudin expression profiles refine decisions around anti‑angiogenic agents, barrier‑modulating therapies, and inclusion in clinical protocols that target tumor tight‑junction biology, with serial cfDNA and microRNA‑21 tracking to evaluate response and minimal residual disease. Infectious disease management integrates H. pylori virulence genotyping, including cagA EPIYA motif diversity, vacA status, and inflammatory gene expression patterns, into eradication strategy design, with antibiotic selection informed by regional sensitivity profiles and host inflammatory tone to minimize resistance and recurrence. In sepsis and septic shock, endothelial and NET‑related biomarkers guide timely deployment of endothelial‑protective measures, anticoagulation strategies when clinically appropriate, and immunomodulation calibrated to molecular risk, while mitochondrial DAMP surveillance shapes de‑escalation and recovery monitoring.

Regenerative and Cellular Therapies: Bridging Organ Failure and Functional Restoration
CLRD’s heritage in hepatic stem/progenitor cell transplantation and bioengineered humanized organs translates into actionable therapies for end‑stage disease. Hepatic stem cells have been deployed to repopulate cirrhotic livers as a supportive modality when transplantation is not immediately feasible, with documented safety and functional gains in decompensated cirrhosis and metabolic disorders such as Crigler–Najjar syndrome. Bioengineered humanized livers and insulin‑producing neo‑organs offer interim support and disease‑modifying potential, accompanied by in vivo tracking of transplanted hepatic progenitors and exosome‑based paracrine strategies to stimulate endogenous repair. Neural precursor cell programs and meningeal scaffold constructs extend the same principles to neuroregeneration, informing protocols for spinal cord injury and neurodegeneration that integrate molecular chaperone and transporter biology to improve graft survival and functional integration.
Integrated Care Pathways: From Molecular Workup to Outcome‑Based Management
Patients enter CLRD’s integrated pathway with a molecular workup that includes genome and epigenome markers, immune transcript signatures, and circulating nucleic acid metrics. The multidisciplinary board maps these results to individualized plans that specify targeted drugs, biologics, metabolic modulators, or regenerative options. Treatment is continuously refined using serial molecular readouts, microRNA‑21 for fibrotic and oncologic surveillance, cfDNA for neurovascular and systemic injury tracking, and immune pathway markers for autoimmune activity, so therapy intensity, combinations, and timing adjust to the patient’s evolving molecular landscape.
Outcomes and Quality: Measuring Impact of Molecularly Guided Care
Outcome frameworks emphasize reduction in time‑to‑diagnosis, fewer therapeutic cycles to achieve remission or response, and improved organ function in regenerative cohorts. For hypertension and cardiometabolic disease, alignment of AT1R genotypes with pharmacotherapy targets better blood pressure control and biochemical normalization. In IBD and autoimmune care, the integration of DNA repair polymorphisms, complement lectin metrics, and inflammatory microRNAs correlates with earlier mucosal healing and reduced steroid exposure. In sepsis, endothelial dysfunction markers and NET surrogates enable stratified care that reduces ICU complications and supports organ recovery trajectories tracked by mitochondrial DAMPs. Regenerative programs report functional bridging and survival benefits while decreasing reliance on systemic immunosuppression through scaffold design and local immune modulation.
What Patients and Clinicians Can Expect at CLRD
The CLRD model emphasizes a seamless experience: molecular sampling with rapid turnaround; clear interpretation that links findings to therapeutic choices; access to targeted drugs, biologics, and cell‑based interventions; and a data‑driven follow‑up cadence anchored in non‑invasive biomarkers. As the portfolio widens, from DNA repair and complement genetics to cfDNA, microRNAs, and engineered tissue supports, patients benefit from care that is demonstrably adaptive and personalized, while clinicians gain decision confidence rooted in reproducible molecular and immunological evidence.
